
KeyPathwayMiner: detecting case-specific biological pathways using expression data. Discovering regulatory and signalling circuits in molecular interaction networks. OmicsAnalyzer: a Cytoscape plug-in suite for modeling omics data. NetAtlas: a Cytoscape plugin to examine signaling networks based on tissue gene expression. VistaClara: an expression browser plug-in for Cytoscape. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BioMart Central Portal-unified access to biological data. WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. Merico, D., Isserlin, R., Stueker, O., Emili, A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. PiNGO: a Cytoscape plugin to find candidate genes in biological networks. Gene ontology: tool for the unification of biology. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles.
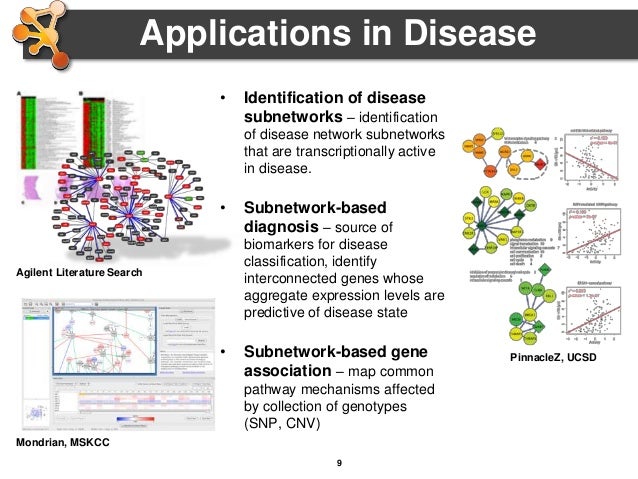
Which clustering algorithm is better for predicting protein complexes? BMC Res. Computational approaches for detecting protein complexes from protein interaction networks: a survey. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. NeMo: Network Module identification in Cytoscape. An automated method for finding molecular complexes in large protein interaction networks. Clustering by passing messages between data points. An efficient algorithm for large-scale detection of protein families. Analyzing biological network parameters with CentiScaPe. Glioblastoma-specific protein interaction network identifies PP1A and CSK21 as connecting molecules between cell cycle-associated genes. Lethality and centrality in protein networks. Computing topological parameters of biological networks. High-throughput generation, optimization and analysis of genome-scale metabolic models. Modularized learning of genetic interaction networks from biological annotations and mRNA expression data. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Literature-curated protein interaction datasets. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Hamosh, A., Scott, A.F., Amberger, J.S., Bocchini, C.A. An architecture for biological information extraction and representation. Pathway Commons, a web resource for biological pathway data. WikiPathways: pathway editing for the people. The Edinburgh human metabolic network reconstruction and its functional analysis. KEGG: Kyoto Encyclopedia of Genes and Genomes. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. APID2NET: unified interactome graphic analyzer. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Pentchev, K., Ono, K., Herwig, R., Ideker, T. Integrating and annotating the interactome using the MiMI plugin for cytoscape. The BioGRID Interaction Database: 2011 update.
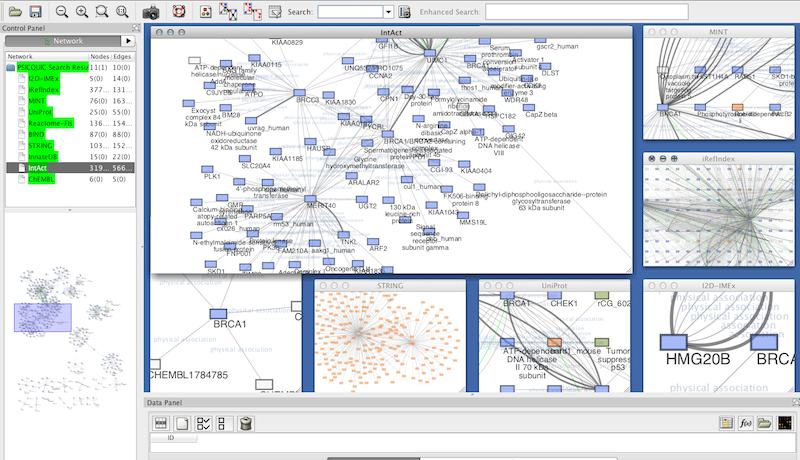
The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. The BioPAX community standard for pathway data sharing. The HUPO PSI's molecular interaction format-a community standard for the representation of protein interaction data. Exploring biological networks with Cytoscape software. Yeung, N., Cline, M.S., Kuchinsky, A., Smoot, M.E.
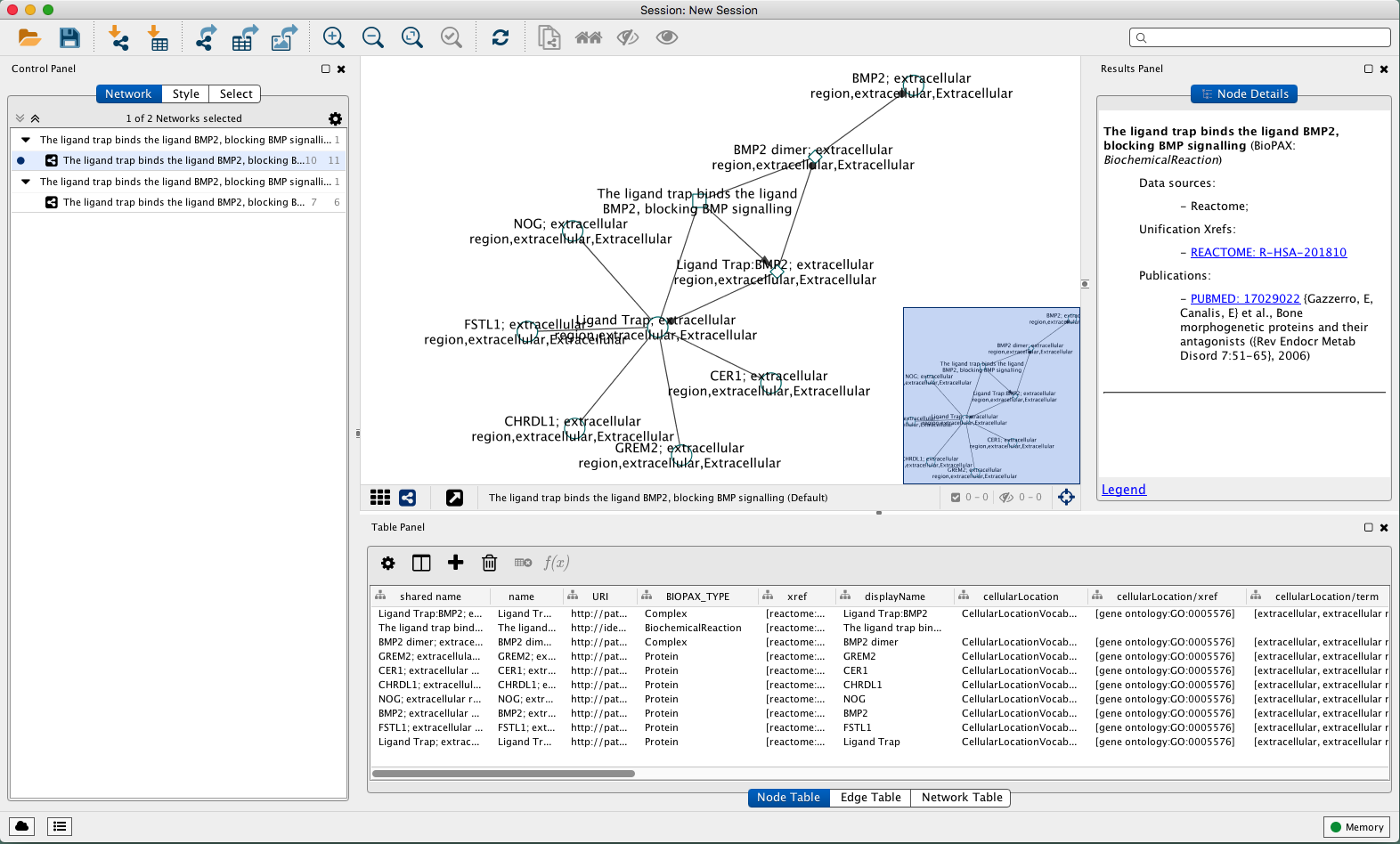
#Cytoscape citation software
Cytoscape: a software environment for integrated models of biomolecular interaction networks. Integration of biological networks and gene expression data using Cytoscape.


 0 kommentar(er)
0 kommentar(er)
